Intragenic two-point recombination was created in order to allow the modification of a particular gene without interfering with the other sub-NNs encoded in other genes. The mechanism of this kind of recombination is exactly the same as in the already familiar two-point recombination, with the difference that the recombination points are chosen within a particular gene.
Consider the following parent chromosomes composed of two genes, each with a Dw domain (Wi,j represents the weights of gene
j in chromosome i):
|
W0,0 = {-0.78, -0.521, -1.224, 1.891, 0.554, 1.237, -0.444, 0.472, 1.012, 0.679}
W0,1 = {-1.553, 1.425, -1.606, -0.487, 1.255, -0.253, -1.91, 1.427, -0.103, -1.625}
0123456789012345601234567890123456
TTababaab14393255QDbabbabb96369304-[0]
Qaabbbabb97872192QDbabbaaa81327963-[1]
W1,0 = {-0.148, 1.83, -0.503, -1.786, 0.313, -0.302, 0.768, -0.947, 1.487, 0.075}
W1,1 = {-0.256, -0.026, 1.874, 1.488, -0.8, -0.804, 0.039, -0.957, 0.462, 1.677} |
Suppose that gene 0 was chosen to recombine and point 1 (between positions 0 and 1) and point 12 (between positions 11 and 12) were chosen as recombination points. Then the following offspring is formed:
|
W0,0 = {-0.78, -0.521, -1.224, 1.891, 0.554, 1.237, -0.444, 0.472, 1.012, 0.679}
W0,1 = {-1.553, 1.425, -1.606, -0.487, 1.255, -0.253, -1.91, 1.427, -0.103, -1.625}
0123456789012345601234567890123456
Taabbbabb97893255QDbabbabb96369304-[0]
QTababaab14372192QDbabbaaa81327963-[1]
W1,0 = {-0.148, 1.83, -0.503, -1.786, 0.313, -0.302, 0.768, -0.947, 1.487, 0.075}
W1,1 = {-0.256, -0.026, 1.874, 1.488, -0.8, -0.804, 0.039, -0.957, 0.462, 1.677} |
Note that the weights of this offspring are exactly the same as the parents. However, due to recombination, the weights expressed in the parents are different from those expressed in the offspring (compare their expressions in
Figure 5.2).

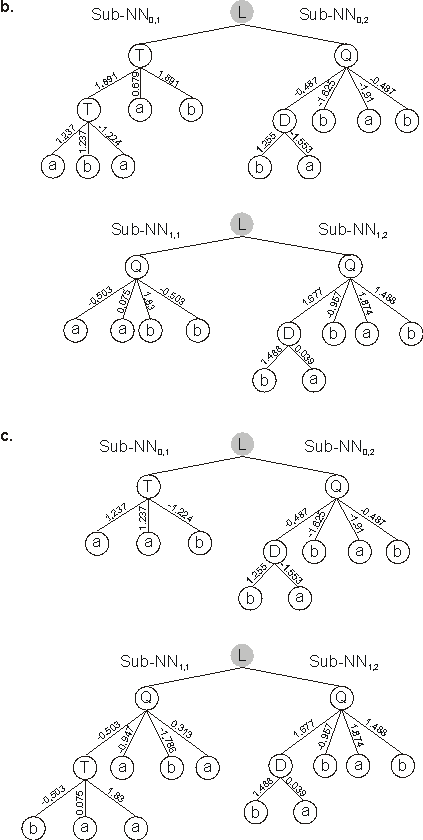
Figure 5.2. Intragenic two-point recombination in multigenic chromosomes encoding neural networks.
a) An event of intragenic two-point recombination illustrated at the chromosome level. Note that the set of weights is not modified by recombination.
b) The sub-NNs codified by the parent chromosomes. c) The sub-NNs codified by the daughter chromosomes. “L” represents a generic linking function.
It is worth emphasizing that this gene-restricted recombination allows a greater control of the modification mechanisms and, consequently, permits a fine tuning of evolution. If we were to use, in multigenic systems, two-point recombination as used in the basic GEA, disrupting chromosomes anywhere, the fine adjustment of the weights in multigenic chromosomes would be an almost impossible task. Restricting two-point recombination to only one gene, however, ensures that only this gene is modified and, consequently, the weights and thresholds of the remaining genes are kept in place.
Remember, however, that intragenic two-point recombination is not the only source of recombination in multigenic neural nets: gene recombination is fully operational in these systems and it can be combined with gene transposition to propel evolution further. And in unigenic systems, the old one-point and two-point recombination are also fully operational as no synchronization of weights is necessary.
|

