Domain specific transposition is restricted to the NN-specific domains – Dw and Dt. Its mechanism, however, is similar to IS transposition. This operator randomly chooses the chromosome, the gene with its respective Dw or Dt, the first position of the transposon, the transposon length, and the target site (also chosen within Dw or Dt). Then it moves the transposon from the place of origin to the target site.
Consider, for instance, the chromosome below with h = 4 (Dw and Dt are shown in different
colors):
|
0123456789012345678901234567890123456
DTQaababaabbaabba05717457362846682867 |
where “T” represents a function of three arguments and “Q” represents a function of four arguments. Suppose that the sequence “84668” was chosen as a transposon and that the insertion site was bond 4 in Dw (between positions
20 and 21). Then the following chromosome is obtained:
|
0123456789012345678901234567890123456
DTQaababaabbaabba05718466874573622867 |
Note that the transposon is deleted at the place of origin, maintaining the domain length.
Suppose that the arrays below represent the weights and the thresholds of both chromosomes:
|
W = {-1.64, -1.834, -0.295, 1.205, -0.807, 0.856, 1.702, -1.026, -0.417, -1.061}
T = {-1.14, 1.177, -1.179, -0.74, 0.393, 1.135, -0.625, 1.643, -0.029, -1.639} |
As their expression shows (Figure 5.1), they encode very different solutions because the weights are moved around and new combinations of weights and thresholds are tested.
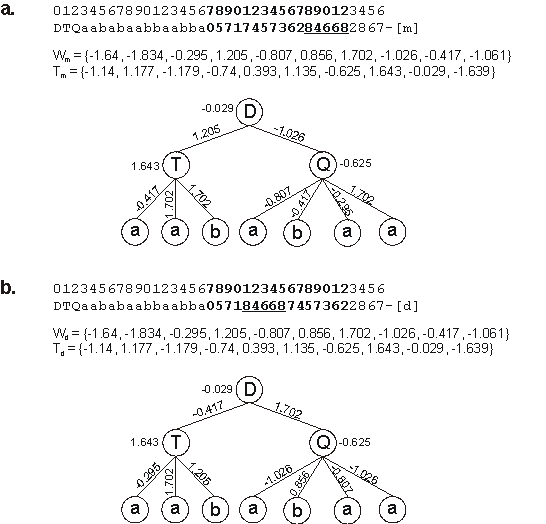
Figure 5.1. Dw-specific transposition. a) The mother neural network.
b) The daughter NN created by Dw-specific transposition. Note that the network architecture is the same for both mother and daughter and that
Wm = Wd and Tm = Td. However, mother and daughter are different because different combinations of weights and thresholds are expressed in these individuals.
|
