|
The incorporation of random numerical constants in Automatically
Defined Functions is also easy and straightforward. As you probably
guessed, the gene structure used to accomplish this includes the
special domain Dc for encoding the random numerical constants,
which, for the sake of simplicity and efficiency, is only
implemented in the genes encoding the ADFs (one can obviously extend
this organization to the homeotic genes, but nothing is gained from
that except a considerable increase in computational effort). The
structure of the homeotic genes remains exactly the same and they
continue to control how often each ADF is called upon and how they
interact with one another.
Consider, for instance, the chromosome with two conventional genes
and their respective arrays of random numerical constants:
|
0123456789001234567890012345678012345678 |
|
|
**?b?aa4701+/Q?ba?8536*0Q-10010/Q-+01111 |
(13) |
|
|
|
|
A0 = {0.664, 1.703, 1.958, 1.178,
1.258, 2.903, 2.677, 1.761, 0.923, 0.796} |
|
|
A1 = {0.588, 2.114, 0.510, 2.359,
1.355, 0.186, 0.070, 2.620, 2.374, 1.710} |
|
The genes encoding the ADFs are expressed exactly as normal genes
with a Dc domain and, therefore, their respective ADFs will, most
probably, include random numerical constants (Figure
10). Then these ADFs with random numerical constants are called
upon as many times as necessary from any of the main programs
encoded in the homeotic genes. As you can see in
Figure 10, ADF0 is invoked twice in Cell0
and once in Cell1, whereas ADF1 is used just
once in Cell0 and called three different times in Cell1.
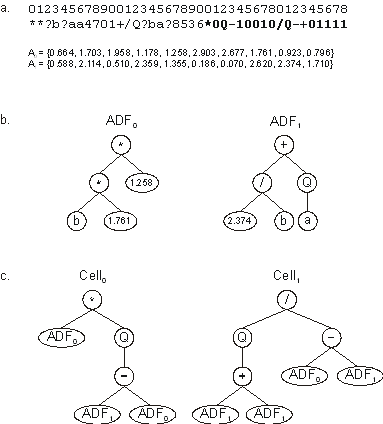
Figure 10. Expression of a multicellular system with
Automatically Defined Functions containing random numerical
constants. a) The chromosome composed of two conventional
genes and two homeotic genes (shown in bold). b) The ADFs
codified by each conventional gene. c) Two different programs
expressed in two different cells.
|
