|
The chromosomes of gene expression programming are usually composed of more than one gene of equal length. For each problem or run, the number of genes, as well as the length of the head, are a priori chosen. Each gene codes for a sub-ET and the sub-ETs interact with one another forming a more complex multi-subunit expression tree.
Consider, for example, the following chromosome with length 39, composed of three genes, each with length 13 (the tails are shown in
blue):
|
012345678901201234567890120123456789012 |
|
|
*Qb+*/bbbabab-a+Qbabbababa/ba-/*bbaaaaa |
(8) |
It has three open reading frames, and each ORF codes for a sub-ET
(Figure 6). The start of each ORF is always given by position 0; the end of each ORF, though, is only evident upon construction of the corresponding sub-ET. As shown in
Figure 6, the first open reading frame ends at position 9; the second ORF ends at position 5; and the last ORF ends at position 2. Thus, GEP chromosomes contain several ORFs of different sizes, each ORF coding for a structurally and functionally unique sub-ET. Depending on the problem at hand, these sub-ETs may be selected individually depending on their respective outputs, or they may form a more complex, multi-subunit expression tree and be selected as a whole. In these multi-subunit structures, individual sub-ETs interact with one another by a particular kind of posttranslational interaction or linking. For instance, algebraic sub-ETs can be linked by addition or multiplication whereas Boolean sub-ETs can be linked by OR, AND or IF.
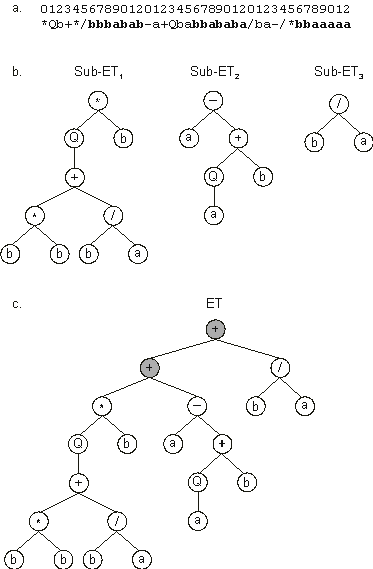
Figure 6. Expression of GEP genes as sub-ETs. a) A three-genic chromosome with the tails shown in bold. Position zero marks the start of each gene.
b) The sub-ETs codified by each gene, which correspond respectively to
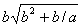 , ,
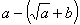 , and , and
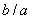 .
c) The result of posttranlational linking with addition, which obviously corresponds to .
c) The result of posttranlational linking with addition, which obviously corresponds to
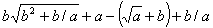 . The linking functions are shown in gray. . The linking functions are shown in gray.
The linking of three sub-ETs by addition is illustrated in
Figure 6, c. Note that the final ET could be linearly encoded as the following K-expression:
|
012345678901234567890 |
|
|
++/*-baQba++Qb*/abbba |
(9) |
However, the use of multigenic chromosomes is more appropriate to evolve solutions to complex problems, for they permit the modular construction of complex, hierarchical structures, where each gene codes for a smaller and simpler building block. These smaller building blocks are separated from each other, and thus can evolve independently. Not surprisingly, these multigenic systems are much more efficient than unigenic ones
(Ferreira 2001, 2002a).
|
