Of the operators with intrinsic modification power, mutation is the most efficient (see
chapter 7). With mutation, populations of individuals adapt very efficiently, allowing the evolution of good solutions to virtually all problems. Typically, I use a mutation rate
pm equivalent to two one-point mutations per chromosome. Considering the length of GEP genomes, this mutation rate is much higher than the mutation rates found in nature (see, e.g.,
Futuyma 1998). Indeed, thanks to elitism, we can have GEP populations undergoing excessive mutation and, nevertheless, evolving very efficiently. As a comparison, the human genome is about 6 X
109 base pairs long and only about 120 new mutations are introduced per genome per generation.
In gene expression programming, mutations can occur anywhere in the chromosome. However, the structural organization of chromosomes must be preserved. Thus, in the heads, any symbol can change into another (function or terminal); in the tails, terminals can only change into terminals. This way, the structural organization of chromosomes is maintained, and all the new individuals produced by mutation are structurally correct programs.
It is worth emphasizing that GEP point mutation is totally unconstrained. This means that, in the heads, functions can be replaced by other functions without concern for the number of arguments each one takes; functions can also be replaced by terminals and vice versa; and obviously terminals can also be replaced by other terminals. Indeed, no restrictions exist whatsoever and, therefore, mutation can be completely exploited to roam freely the fitness landscape.
The workings of mutation can be analyzed in Figure
3.11. For this analysis, only mutation was used as the source of genetic variation. The mutation rate was as usual equivalent to two one-point mutations per chromosome, which, in this case, gives
pm = 0.143. The populations shown in Figure
3.11 were obtained in a run to solve the already familiar Majority
(a, b, c) function problem. Note that chromosome 1 of generation 6 encodes a perfect solution to the majority function and, therefore, has maximum fitness.
|
Generation N: 0
01234560123456
NabbabbAAccbcb-[0] = 3
NAabbcaNbbbcca-[1] = 2
OcOcaaaNaOabaa-[2] = 4
AaAcccbAbccbbc-[3] = 7
AObbabaAOcaabc-[4] = 7
AAAbaacONOaabc-[5] = 4
AAccbcaNNcbbac-[6] = 6
NOccabaOcbabcc-[7] = 4
NOAcbbbAaNabca-[8] = 2
NacbbacAbccbbc-[9] = 3
...
Generation N: 5
Generation N: 6
01234560123456
01234560123456
AabbabcAOcaabc-[0] = 7
AOAbabacbcaaba-[0] = 7
babbabcAOcaabc-[1] = 7 AabbabcAOcbabc-[1] = 8
AOAacbcOAOcaac-[2] = 6 AabbabcAccaabc-[2] = 7
ANbbabcAOcaabc-[3] = 6 NAAbaacONaaacc-[3] = 4
AOAbabacbcaaba-[4] = 7 AOAbabacbcaaba-[4] = 7
AabcaccAONaabc-[5] = 6 AabbbbcAONaabb-[5] = 6
AOAccbaAbaabbc-[6] = 6 AOAbabacNcabba-[7] = 7
AObcabaAbNcaba-[7] = 6 AOAccbaAbaabbc-[6] = 6
NAAbbacONOacca-[8] = 3 NAAbbacONOacaa-[8] = 4
AONbabacbcaaba-[9] = 5 AObbabaAAcaabc-[9] = 7
|
Figure 3.11. An initial population and its later descendants created, via mutation, to solve the Majority
(a, b, c) function problem. The chromosomes encode sub-ETs linked by OR. Note that none of the later descendants are identical to their ancestors of generation 0. The perfect solution found in generation 6 (chromosome 1) and one of its putative ancestors (chromosome 0 of generation 5) are shown in
blue. Note that chromosomes 1 and 3 of generation 5 are also good candidates to be the predecessors of the perfect solution. In both cases, two point mutations would have occurred during reproduction.
The most probable ancestors of this perfect solution are chromosomes 0, 1, and 3 of generation 5. In the first case, only one point mutation would have occurred during reproduction whereas two point mutations would have occurred in the last two cases.
Figure 3.12 compares the ETs of one of these putative ancestors (chromosome 3) with the daughter ET, i.e., before and after mutation. In this case, two point mutations occurred during reproduction of chromosome 3: one changed the “N” at position 1 in gene 1 into “a”; and another changed the “a” at position 3 in gene 2 into “b”. Note that the first mutation changed significantly the
sub-ET1, shortening the original sub-ET in one node. Note also that, although the second mutation did not change the shape of the sub-ET, the expression encoded in this sub-ET is not the same.
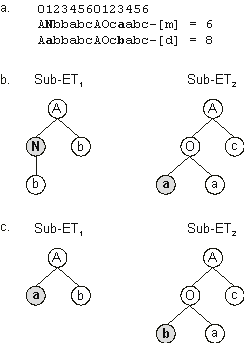
Figure 3.12. Illustration of mutation. a) The mother and daughter chromosomes with the mutation points shown in bold.
b) The sub-ETs encoded by the mother chromosome (before mutation).
c) The sub-ETs encoded by the daughter chromosome (after mutation). The mutation nodes are shown in gray. Note that the first mutation changed significantly the
sub-ET1, shortening the original sub-ET in one node.
Let us analyze more closely the populations shown in Figure
3.11. On the one hand, we can see that several mutations have a neutral effect. For instance, chromosome 7 of generation 6 is a descendant of chromosome 4 of generation 5. These chromosomes differ only at positions 1 and 4 in gene 2. The expression of these chromosomes gives identical ETs
(Figure 3.13), as the mutations occurred downstream of the termination point of
ORF2. As we have seen, mutations occurring in the non-coding region of a gene have a neutral effect, as they have no expression in the “organism” itself.
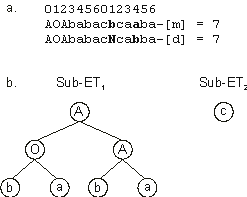
Figure 3.13. Illustration of neutral mutations. a) The mother and daughter chromosomes with the mutation points shown in bold.
b) The sub-ETs encoded by both chromosomes. Note that the mutations shown in (a) have no expression in the final ET as they occurred downstream of the termination point.
On the other hand, we can see that, in GEP, mutations in the coding sequence of a gene have usually a very profound effect: most of the times they reshape the ET drastically. However, this capability to reshape profoundly the ET is fundamental for evolvability (see
chapter 7 for a comparison of the genetic operators). Indeed, the results presented throughout this book clearly show that our too human wish to keep intact the small functional blocks and recombine them carefully (as is done in GP) is conservative and works poorly. Genotype/phenotype systems can find much more efficient ways of creating and manipulating their own building blocks. These building blocks are very different from the ones a mathematician would have chosen but, nonetheless, they work much more efficiently.
And finally, it is worth emphasizing that in GEP there are no constraints both in the kind of mutation and the number of mutations in a chromosome as, for all cases, the newly created individuals are syntactically correct programs. This important feature distinguishes GEP from all GP-style systems, where point mutations would have resulted most of the times in invalid programs. And an evolutionary algorithm unable to use this powerful operator is severely restricted, since mutation is the most important agent of genetic diversification (see
chapter 7).
|

