The linking of sub-ETS by a particular linking function is simple and efficient in evolutionary terms. Indeed, from unigenic to multigenic systems, efficiency increases considerably. Despite this artificial increase in complexity (“artificial” in the sense that it was not evolved by the system itself), evolution in multigenic systems still occurs efficiently and therefore they can be efficiently used to evolve solutions to different problems. In principle, it is possible to impose from outside higher levels of complexity, but this is no guarantee that an increase in performance will be obtained. Evolutionary complex systems are not created this way: higher levels of complexity are created above lower levels and the evolution of complexity occurs more or less continuously. Notwithstanding, the interaction of sub-ETs in GEP can be programmed using a higher level of complexity. For that purpose a special class of genes was created – homeotic genes – which control the development of the individual. The expression of such genes results in different programs or cells. Thus, homeotic genes determine which genes are expressed in which cell and how they interact with one another.
So, what is the structure of homeotic genes? They have exactly the same kind of structure as conventional genes and are built using an identical process. The heads contain, in this case, linking functions and a special class of terminals – genic terminals – representing conventional genes. The tails contain obviously only genic terminals.
Consider, for instance, the following chromosome:
| 01234560123456012345601234567890 |
|
|
*-*aaaa*a/aaaa+/+aaaa+*2+Q322122 |
(2.26) |
It codes for three conventional genes and one homeotic gene (shown in
blue). The conventional genes code as usual for three different sub-ETs. The homeotic gene controls the interactions between the different sub-ETs. Note that homeotic genes have their specific length and their specific set of functions. In this particular case, the head length of the homeotic gene
hH is equal to five whereas for the other genes the head length is equal to three; the function set of the homeotic gene
FH is equal to FH = {+, *, Q} whereas for the other genes the function set consists of
F = {+, -, *, /}. As shown in Figure
2.12, this kind of representation not only allows the evolution of linking functions but also allows code reuse. As you probably noticed, this is an extremely elegant and simple form of generating in GEP what is known as automatically defined functions (ADFs). Indeed, any conventional gene in this cellular representation can not only be used as many times as necessary but also establish different interactions with the other genes.
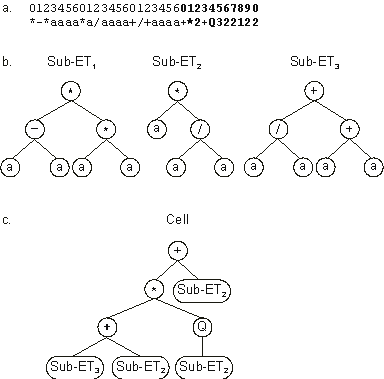
Figure 2.12. Expression of chromosomes containing a single homeotic gene.
a) The chromosome composed of three conventional genes and one homeotic gene (shown in bold).
b) The sub-ETs codified by each conventional gene. c) The final program or cell. Note that this cellular system allows code reuse as each conventional gene codes for an automatically defined function.
The use of more than one homeotic gene results obviously in a multicellular system where each homeotic gene puts together a different consortium of genes. Consider, for instance, the following chromosome:
| 01234560123456012345601234560123456 |
|
|
/a/aaaa+a*aaaa*++aaaaQ*21122+/31333 |
(2.27) |
It codes for three conventional genes and two homeotic genes (shown in
blue). Its expression (Figure 2.13) results in two different cells or programs, each expressing different genes in different ways.
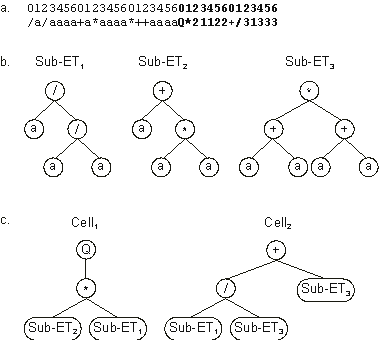
Figure 2.13. Expression of chromosomes containing multiple homeotic genes.
a) The chromosome composed of three conventional genes and two homeotic genes (shown in bold).
b) The sub-ETs codified by each conventional gene. c) Two different programs expressed in two different cells. Note how different cells put together different consortiums of genes.
|

