As we have seen in the examples above, although a very simple system, gene expression programming exhibits already a complex development. The most complex individuals evolved by GEP contain, besides the head and the tail, extra domains encoding one-element sub-ETs. These one-element sub-ETs interact with the main sub-ET encoded in the head/tail domain, forming a more complex entity with a complex network of interactions.
One such architecture was developed to manipulate random numerical constants in symbolic regression
(Ferreira 2001). For instance, the following chromosome:
| 01234567890123456789012 |
|
|
+*?+?*+a??aaa??09081345 |
(2.28) |
contains an extra domain Dc (shown in blue) encoding random numerical constants. Its translation is shown in
Figure 2.14. In section 4.2 we will learn how these sub-ETs interact with one another so that the individual is fully developed.
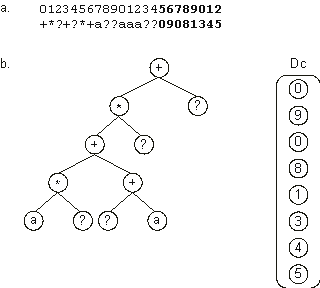
Figure 2.14. Translation of chromosomes with multiple domains.
a) A multi-domain chromosome composed of a conventional head/tail domain and an extra domain (Dc) encoding random numerical constants (shown in bold).
b) The sub-ETs codified by each gene. The one-element sub-ETs encoded in Dc are placed together apart. “?” represents the random numerical constants encoded in the numerals of Dc. How all these sub-ETs interact will be shown in
section 4.2.
Multiple domains are also used to design neural networks totally encoded in a linear genome (see
chapter 5). These neural networks are one of the most complex kind of individual evolved by GEP. In this case, the network architecture is encoded in a conventional head/tail domain whereas the weights and thresholds are encoded in two extra domains, Dw and Dt, each composed of one-element genes. The chromosome below contains two extra domains encoding the weights and the thresholds (the domains are shown in different
colors):
| 012345678901234567890123456789012345 |
|
|
DUbUTdaedbfaabad42998409791482467584 |
(2.29) |
Its translation is shown in Figure 2.15. In
chapter 5 we will learn the rules of their complete development and how populations of these complex individuals evolve, finding solutions to problems in the form of a GEP encoded neural network.
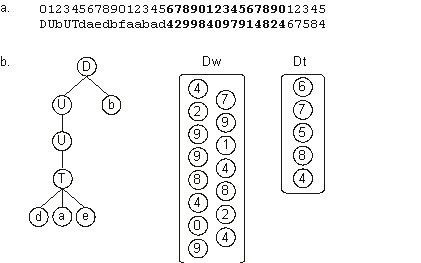
Figure 2.15. Translation of chromosomes with multiple domains.
a) A multi-domain chromosome composed of a conventional head/tail domain encoding the neural network architecture, and two extra domains – one encoding the weights (Dw) of the neural network and another the thresholds (Dt). Dw and Dt are shown in different shades.
b) The sub-ETs codified by each gene. The one-element sub-ETs encoded in Dw and Dt are placed together apart. “U”, “D”, and “T” represent, respectively, functions with connectivity one, two, and three. How all these sub-ETs interact will be shown in
chapter 5.
|

